NACIA Digital PCR Primer Probe Design
NACIA digital PCR primers are different from common PCR primer design requirements: the purpose of common PCR is to obtain the target gene, which is not strict for non-specific reactions and primer dimers; and digital PCR primers are as non-specific as real-time PCR primers. And the primer dimer is strictly required.
General Principles for NACIA Digital PCR Primers and Probe Design
v Select the probe first, then design the primer as close as possible to the probe.
v The selected column should be highly specific and try to select the amplified fragment with the smallest secondary structure.
v The amplification length should not exceed 400 bp, ideally it can be within 50-150 bp.
v Keep the GC content between 30% and 80%.
v To ensure efficiency and repeatability, repeated nucleotide sequences should be avoided, especially G (no 4 consecutive Gs).
v Pair and probe the primers and probes to avoid the formation of dimers and hairpin structures.
v Some factors that need additional consideration:
☆ The base 3 is avoided at the 3' end of the primer, and 3 or more consecutive identical bases are avoided at the 3' end of the primer.
☆ If the secondary structure cannot be avoided, increase the annealing temperature accordingly.
☆ Try to avoid the Dimer (dimer) matching area at the 3' end.
☆ Try to avoid several consecutive base mismatches at the 3' end to form False priming
☆ Measure the stability of the secondary structure. If the free energy value is greater than 0, the structure is unstable and will not interfere with the reaction. If the free energy value is less than 0, the structure can interfere with the reaction and try to ensure the absolute value of free energy <10 (optional).
In order to avoid the detection of DNA during gene expression, the design should be as far as possible across the intron region, which ensures that the amplified product can only use mRNA as a template.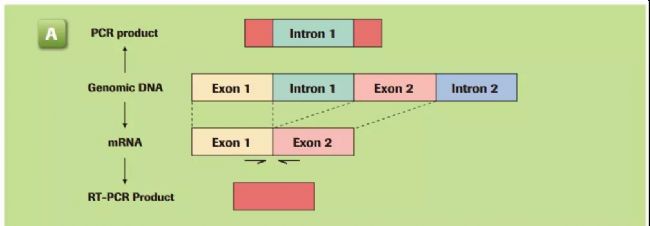
Digital PCR
☆ Tm value: 58 ° C to 60 ° C, TM difference between primers avoids more than 2 ° C
☆ GC content: 30% to 80%
☆ There should be no more than 2 G+Cs in the last 5 bases of the 3' end
☆ Try to avoid repeated nucleotide sequences, especially 4 consecutive G
☆ The probe position is as close as possible to the upstream primer, but it cannot overlap
Basic principles of NACIA digital PCR probe design ☆ Probe length: 13-25 bases (MGB probe); 13-30 bases (ordinary Taqman probe)
☆ Tm value: 68 °C to 70 °C (PrimerExpress calculation results); usually 5-10 °C higher than the primer TM value (at least 5 °C)
☆ GC content: 30% to 80%
☆ The first base at the 5' end cannot be G
☆ Try to avoid repeated nucleotide sequences, especially 4 consecutive G; avoid 6 consecutive A occurrences ☆ For MGB probes, try to avoid 2 or more consecutive Cs in the middle of the probe The base at the 3' end should be avoided as "GGG" or "GGAG"
☆ For FAM-labeled probes, the second base at the 5' end cannot be G
☆ In the entire probe, the content of base C is significantly higher than the content of G.
NACIA Digital PCR Probe Fluorescent Label Selection: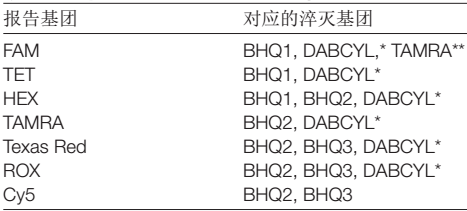
☆ Probes for different target genes label different fluorophores, such as FAM, HEX or CY5, and select the fluorescent reporter group with the least penetration between the emission wavelengths.
☆ Also compatible with the fluorescence detection channel parameters of the digital PCR system.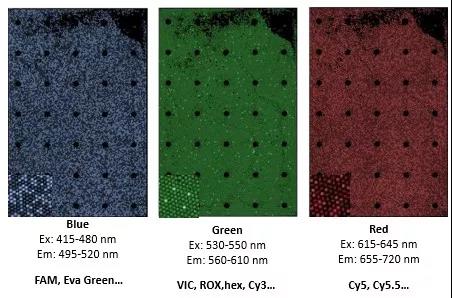
General Principles for NACIA Digital PCR Primers and Probe Design
v Select the probe first, then design the primer as close as possible to the probe.
v The selected column should be highly specific and try to select the amplified fragment with the smallest secondary structure.
v The amplification length should not exceed 400 bp, ideally it can be within 50-150 bp.
v Keep the GC content between 30% and 80%.
v To ensure efficiency and repeatability, repeated nucleotide sequences should be avoided, especially G (no 4 consecutive Gs).
v Pair and probe the primers and probes to avoid the formation of dimers and hairpin structures.
v Some factors that need additional consideration:
☆ The base 3 is avoided at the 3' end of the primer, and 3 or more consecutive identical bases are avoided at the 3' end of the primer.
☆ If the secondary structure cannot be avoided, increase the annealing temperature accordingly.
☆ Try to avoid the Dimer (dimer) matching area at the 3' end.
☆ Try to avoid several consecutive base mismatches at the 3' end to form False priming
☆ Measure the stability of the secondary structure. If the free energy value is greater than 0, the structure is unstable and will not interfere with the reaction. If the free energy value is less than 0, the structure can interfere with the reaction and try to ensure the absolute value of free energy <10 (optional).
In order to avoid the detection of DNA during gene expression, the design should be as far as possible across the intron region, which ensures that the amplified product can only use mRNA as a template.

Digital PCR

Digital PCR
NACIA Digital PCR Primer Design Basic Principles ☆ Amplified fragment length: 50-150 bases ☆ Primer length: 18-24 bases (20 bases optimal) ☆ Tm value: 58 ° C to 60 ° C, TM difference between primers avoids more than 2 ° C
☆ GC content: 30% to 80%
☆ There should be no more than 2 G+Cs in the last 5 bases of the 3' end
☆ Try to avoid repeated nucleotide sequences, especially 4 consecutive G
☆ The probe position is as close as possible to the upstream primer, but it cannot overlap
Basic principles of NACIA digital PCR probe design ☆ Probe length: 13-25 bases (MGB probe); 13-30 bases (ordinary Taqman probe)
☆ Tm value: 68 °C to 70 °C (PrimerExpress calculation results); usually 5-10 °C higher than the primer TM value (at least 5 °C)
☆ GC content: 30% to 80%
☆ The first base at the 5' end cannot be G
☆ Try to avoid repeated nucleotide sequences, especially 4 consecutive G; avoid 6 consecutive A occurrences ☆ For MGB probes, try to avoid 2 or more consecutive Cs in the middle of the probe The base at the 3' end should be avoided as "GGG" or "GGAG"
☆ For FAM-labeled probes, the second base at the 5' end cannot be G
☆ In the entire probe, the content of base C is significantly higher than the content of G.
NACIA Digital PCR Probe Fluorescent Label Selection:

NACIA Digital PCR
NACIA Digital PCR Multiple Probe Design Principles ☆ The Tm values ​​of the primers or probes of all target genes should be similar. The Tm value of the primer is recommended to be 55-60 ° C, and the Tm value of the probe is higher than the primer Tm value 5-10 ° C. ☆ Probes for different target genes label different fluorophores, such as FAM, HEX or CY5, and select the fluorescent reporter group with the least penetration between the emission wavelengths.
☆ Also compatible with the fluorescence detection channel parameters of the digital PCR system.

NACIA Digital PCR
NACIA Digital PCR recommends primer probe design software: Beacon designer, Primer 3, Primer express, etc. contact us:
Beijing Deep Blue Cloud Biotechnology Co., Ltd.
Address: Room 201, 2nd Floor, Unit 2, Building 2, No. 25, Jinghai 4th Road, Beijing Economic and Technological Development Zone, Beijing
phone
mailbox:
Official website:
Children Face Mask,Kids Medical Disposable Face Mask,Kids 3 Ply Disposable Face Mask,Kids Nonwoven Disposable Face Mask
KUTA TECHNOLOGY INDUSTRY CO.,LIMITED , https://www.kutasureblue.com